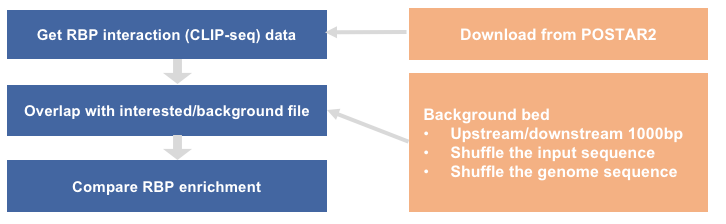
workflow
1. get CLIP-seq data
1.1 download from http://lulab.life.tsinghua.edu.cn/postar/download.php
1.2 get from /BioII/lulab_b/shared/projects/POSTAR2/CLIP_seq/bed
human_RBP_eCLIP_hg38.txt
chr1 187044 187087 human_RBP_eCLIP_ENCODE_1 0 - AARS eCLIP K562 ENCODE 3.02982873699729
chr1 267418 267466 human_RBP_eCLIP_ENCODE_2 0 - AARS eCLIP K562 ENCODE 5.50800008227284
chr1 630751 630792 human_RBP_eCLIP_ENCODE_3 0 - AARS eCLIP K562 ENCODE 3.5454755693998
explanation for each column
column1:chromosome
column2:peak start
column3:peak end
column4:name
column5:0
column6:strand
column7:RBP name
column8:CLIP-seq technology, peak calling method
column9:cell line or tissue
column10:data accession
column11:score(Piranha score: Peak heights from the CLIP-seq data; PARalyzer score: T-to-C transition ratios ranging from 0 to 1, ratios greater than 0.5 indicate protein-binding while less than 0.5 indicate without protein-binding; CIMS score: Mismatch peak heights from the CLIP-seq data; CITS score: Truncated peak heights from the CLIP-seq data; eCLIP score: -log10 P-value, the threshold is 3.)
2. prepare interested bed and background bed
2.1 interested bed
up.bed
chr1 1167104 1167134 ENSG00000207730.3__1167104__1167134__1 . +
chr1 1167119 1167149 ENSG00000207730.3__1167119__1167149__2 . +
chr1 1167134 1167164 ENSG00000207730.3__1167134__1167164__3 . +
2.2 prepare background bed from interested bed
the same as method in sequence_motif (1) downstream/upstream 1000bp (2) shuffle the input sequence (3) shuffle the genome sequence
3. overlap between CLIP-seq peaks and interested/background bed
use intersectBed (bedtools) command to get the overlaps of CLIP-seq peaks
intersectBed -wa -wb -s -a up.bed -b human_RBP_eCLIP_hg38.txt >up.eCLIP.hg38.txt
intersectBed -wa -wb -s -a up_background.bed -b human_RBP_eCLIP_hg38.txt >up_background.eCLIP.hg38.txt
4. compare RBP enrichment
4.1 percentage of region with CLIP-seq peaks
cut -f 4 up.bed |sort -u|wc -l
1685
cut -f 4 up.eCLIP.hg38.txt |sort -u|wc -l
663
cut -f 4 up_background.bed |sort -u|wc -l
1685
cut -f 4 up_background.eCLIP.hg38.txt |sort -u|wc -l
544
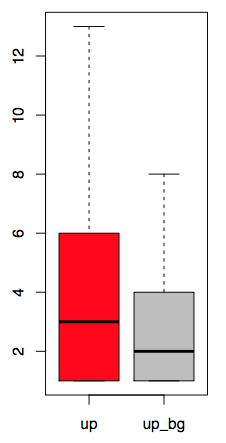
4.2 RBP enrichment in each region
cat <(cut -f 4,13 up.eCLIP.hg38.txt |sort -u|cut -f 1|sort|uniq -c|sed 's/^[ \t]*//g'|sed 's/ /\t/g'|awk 'BEGIN{FS="\t";OFS="\t"}{print $1,$2,"up"}') <(cut -f 4,13 up_background.eCLIP.hg38.txt |sort -u|cut -f 1|sort|uniq -c|sed 's/^[ \t]*//g'|sed 's/ /\t/g'|awk 'BEGIN{FS="\t";OFS="\t"}{print $1,$2,"up_bg"}')|sed '1i density\tname\tclass' >RBP_enrichment.txt
screen short of “RBP_enrichment.txt”
density name class
1 ENSG00000004897.11__47141883__47141913__96 up
2 ENSG00000104723.20__15540418__15540448__14 up
2 ENSG00000115758.12__10441554__10441584__100 up
boxplot of RBP enrichment