structure motif analysis
1) workflow
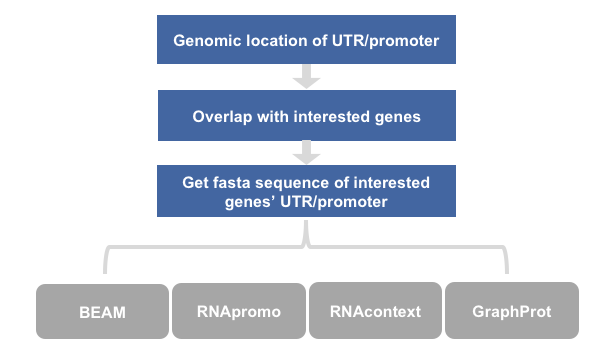
如果你学习完了Sequence Motif,那么本节所需文件就在你的docker bioinfo_tsinghua容器的/home/test/motif/structure_motif/目录下,如果没学上面的小节,你也可以按照这个目录自己放置文件,源文件在这里清华大学云盘文件名为example_motif.tar.gz
2) running steps
BEAM与RNAfold的安装配置
root下,bioinfo_tsinghua container
清华大学云盘下载BEAREncoder.tar.gz放在share文件夹中
# BEAM的配置,待完善
docker exec -it -u root bioinfo_tsinghua bash
mkdir /home/test/software/BEAM
cd /home/test/software/BEAM
wget https://github.com/noise42/beam/archive/v2.0.tar.gz
tar -zxvf v2.0.tar.gz
cd beam-2.0/
cp ~/share/BearEncoder.new.jar ./
# RNAfold的配置
mkdir /home/test/software/ViennaRNA
wget https://www.tbi.univie.ac.at/RNA/download/sourcecode/2_4_x/ViennaRNA-2.4.14.tar.gz
tar -zxvf ViennaRNA-2.4.14.tar.gz
cd ViennaRNA-2.4.14
mkdir /home/test/software/ViennaRNA/ViennaRNA
./configure --prefix=/home/test/software/ViennaRNA/ViennaRNA
make
make install
(1) get interested sequence and control sequence as sequence motif analysis
1.1 BEAM
http://beam.uniroma2.it/home
test身份进入容器
# 在这个路径下已经准备了练习文件
/home/test/motif/structure_motif/BEAM
cd /home/test/motif/structure_motif/BEAM
1.2 Use RNAfold to get dot-bracket
Compute the best (MFE) structure for this sequence (primary sequence with dot-bracket)
RNAfold <test.fa >dot.fa
less dot.fa
# 查看生成的序列及点括号文件dot.fa
1.3 Get file with BEAR notation —> fastB (fastBEAR).
awk '/^>/ {print; getline; print; getline; print $1}' dot.fa >dot_to_encode.fa
java -jar /home/test/software/BEAM/beam-2.0/BearEncoder.new.jar dot_to_encode.fa BEAMready.fa
1.4 get structure motifs
java -jar /home/test/software/BEAM/beam-2.0/BEAM_release_1.5.1.jar -f BEAMready.fa -w 10 -W 40 -M 3

1.5 visualize motifs with weblogo
1.5.1 install weblogo
install weblogo root身份登录容器
pip install weblogo
1.5.2 visualize structure motifs
test用户
cd /home/test/motif/structure_motif/BEAM/risultati/BEAMready/webLogoOut/motifs
weblogo -a 'ZAQXSWCDEVFRBGTNHY' -f BEAMready_m1_run1_wl.fa -D fasta \
-o out.jpeg -F jpeg --composition="none" \
-C red ZAQ 'Stem' -C blue XSW 'Loop' -C forestgreen CDE 'InternalLoop' \
-C orange VFR 'StemBranch' -C DarkOrange B 'Bulge' \
-C lime G 'BulgeBranch' -C purple T 'Branching' \
-C limegreen NHY 'InternalLoopBranch'
cp -p out.jpeg ~/share/
# 进入share文件夹查看输出结果
1.5.3 example output

3) other tools
3.1 RNApromo
https://genie.weizmann.ac.il/pubs/rnamotifs08/64bit_exe_rnamotifs08_motif_finder.tar.gz
3.2 GraphProt:modelling binding preferences of RNA-binding proteins
https://github.com/dmaticzka/GraphProt
3.3 RNAcontext: A New Method for Learning the Sequence and Structure Binding Preferences of RNA-Binding Proteins
http://www.cs.toronto.edu/~hilal/rnacontext/