structure-seq analysis (Shape-map)
background
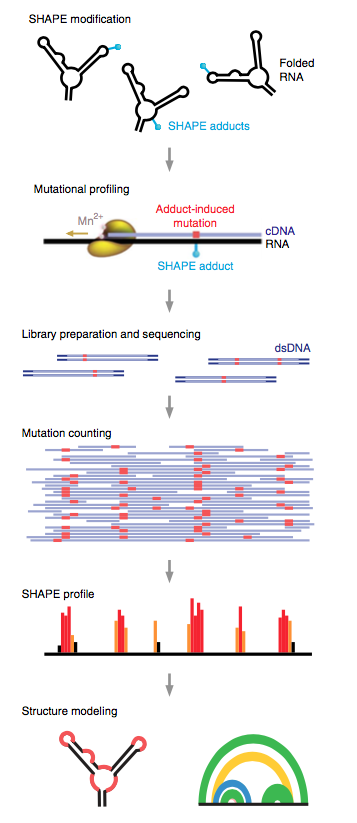
RNA is treated with a SHAPE reagent that reacts at conformationally dynamic nucleotides. During reverse transcription, polymerase reads through chemical adducts in the RNA and incorporates a nucleotide noncomplementary to the original sequence (red) into the cDNA. The resulting cDNA is sequenced using any massively parallel approach to create a mutational profile. Sequencing reads are aligned to a reference sequence, and nucleotide-resolution mutation rates are calculated, corrected for background and normalized, producing a standard SHAPE reactivity profile. SHAPE reactivities can then be used to model secondary structures, visualize competing and alternative structures, or quantify any process or function that modulates local nucleotide RNA dynamics.
1.shapemapper
ShapeMapper automates the calculation of RNA structure probing reactivities from mutational profiling (MaP) experiments, in which chemical adducts on RNA are detected as internal mutations in cDNA through reverse transcription and read out by massively parallel sequencing.
shapemapper \
--target TPP.fa \
--name "example-results" \
--overwrite \
--modified --folder TPPplus \
--untreated --folder TPPminus \
--star-aligner
--verbose
#--denatured --folder TPPdenat
input
- TPPplus: modified fasta file
- TPPminus: untreated fasta file
output
- example-results_TPP_histograms.pdf
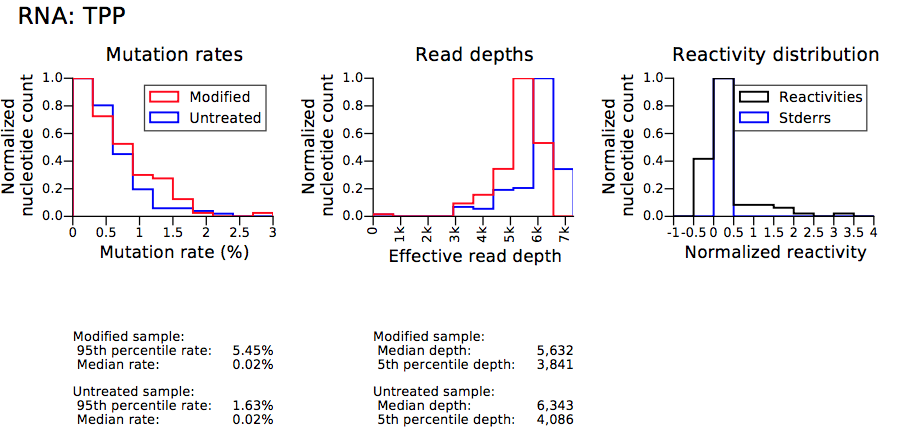
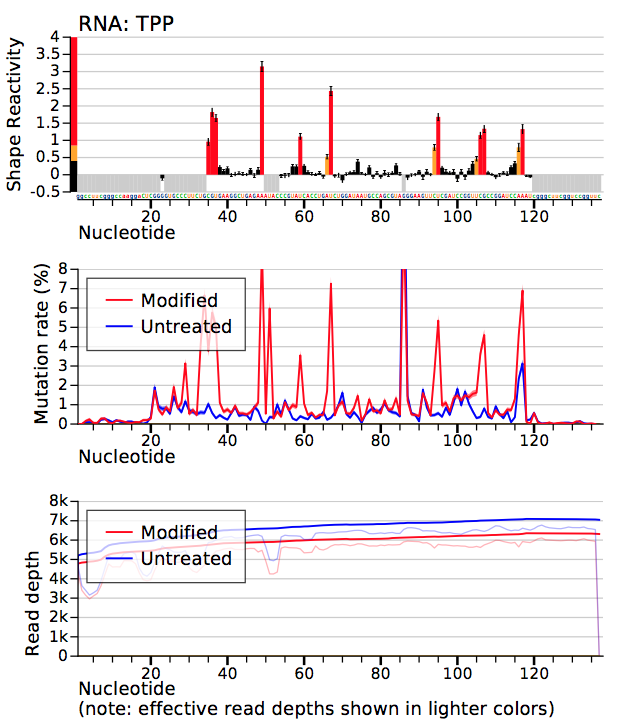
- example-results_TPP_profiles.pdf
- example-results_TPP.shape
1 -999 2 -999 3 -999explanation for each column:
column1: nucleotide number column2: reactivity - example-results_TPP.map
1 -999 0 G 2 -999 0 G 3 -999 0 Cexplanation for each column:
column1: nucleotide number column2: reactivity column3: standard error column4: nucleotide sequence - example-results_TPP_profile.txt
Nucleotide Sequence Modified_mutations Modified_read_depth Modified_effective_depthModified_rate Untreated_mutations Untreated_read_depth Untreated_effective_depth Untreated_rate Denatured_mutations Denatured_read_depth Denatured_effective_depth Denatured_rate Reactivity_profile Std_err HQ_profile HQ_stderr Norm_profile Norm_stderr 1 g 0 4788 4335 0.000000 0 5206 4666 0.000000 0 nan 0.000000 0.000000 nan nan nan nan 2 g 0 4837 3405 0.000000 0 5270 3643 0.000000 0 nan 0.000000 0.000000 nan nan nan nan